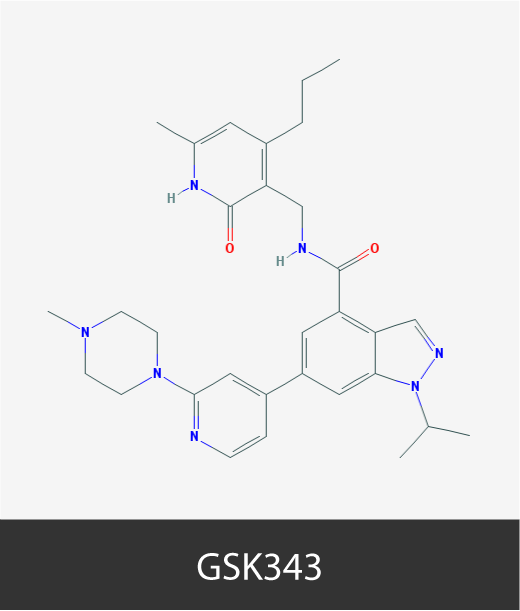
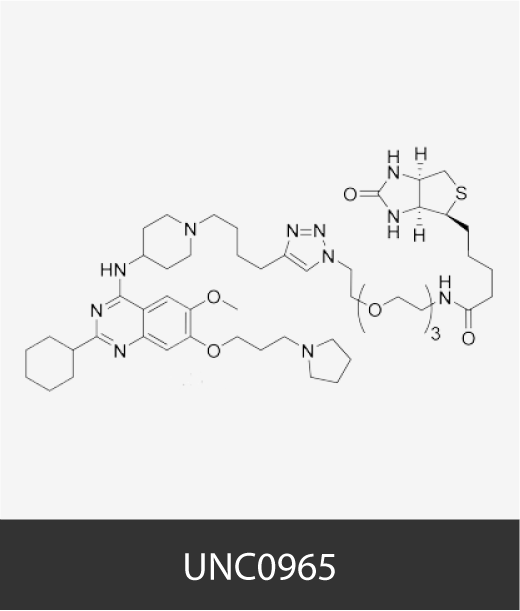
To cite this section of the Cancer Epigenetics Drug Database
Reference: I. Hübscher, M. Wachtel, and I.M. Bennani-Baiti. Histone methyltransferase inhibitors. Cancer Epigenetics Drug Database (CEDD) – Experimental dataset, Cancer Epigenetics Society (https://ces.b2sg.org/cedd/experimental_hmti/), 2019.
Chemical structures of cancer epigenetic drugs
Brief description of cancer epigenetic drugs
Functional assays used to assess the activity of cancer epigenetic drugs
A word about the Cancer Epigenetics Drug Database – HMTi section
Welcome to the Cancer Epigenetic Drug Database – Experimental dataset for histone methyltransferase inhibitors (HMTi). The epigenetics drug database gives an overview of cancer cell lines sensitivity to HMTi in various functional assays (see assays abbreviations below). The page starts off with the chemical structures of the epigenetic drugs, a succinct description of the epigenetic drugs and functional assays used to test them in cancer cell lines, followed by Tables summarizing the data. Some of the epigenetic drugs also have clinical activities which are summarized in this page. Please note that the epigenetic drug solubility values provided here are only to be used as a guide, and that solubility depends on several variables such as the purity of the solute and solvent used, as well as the temperature used. Similarly, the IC50 values provided herein can be highly dependent on the type of functional assay used and length of incubation of cells with the epigenetic drugs.
The Cancer Epigenetics Drug Database is a work in progress and we rely on our members and readers to bring to our attention data that ought to be included here. We, therefore, welcome any information that you may provide that would help us make this database as useful a tool for your research as possible. You may email us your suggestions at info@ces.b2sg.org, Subject line: CEDD – HMTi, Experimental dataset. The Cancer Epigenetics Drug Database is periodically updated, so please check it again for new data that may be relevant to your research.
Notes:
Histone methyltransferase inhibitor (HMTi) activities in cancer cells
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information3 | ID# |
| 22Rv1 | > 10 | 5 | GLO | ||
| Calu-1 | > 10 | 5 | GLO | ||
| Calu-6 | > 10 | 5 | GLO | ||
| Daoy | > 10 | 7 | GLO | ||
| DU145 | > 10 | 5 | GLO | ||
| G-401 | > 10 | 7 | GLO | ||
| HL-60 | > 10 | 5 | GLO | ||
| HT-1080 | > 10 | 5 | GLO | ||
| IMR-32 | > 10 | 12 | GLO | ||
| K-562 | > 10 | 5 | GLO | ||
| Kasumi-1 | > 10 | 5 | GLO | ||
| KMS11 | > 10 | 5 | GLO | ||
| KMS12 | > 10 | 5 | GLO | ||
| L-363 | > 10 | 5 | GLO | ||
| LN-229 | > 10 | 5 | GLO | ||
| LNCap | > 10 | 5 | GLO | ||
| MOLT-16 | > 10 | 5 | GLO | ↓ global H3K9me2, ↑ G9a | |
| MV4-11 | > 10 | 5 | GLO | G0/G1 arrest | |
| NCI-H1975 | > 10 | 5 | GLO | ||
| NCI-H2009 | > 10 | 5 | GLO | ||
| NCI-H358 | > 10 | 5 | GLO | ||
| NCI-H441 | > 10 | 5 | GLO | ||
| NCI-H522 | > 10 | 5 | GLO | ||
| NCI-H661 | > 10 | 5 | GLO | ||
| NCI-H929 | > 10 | 5 | GLO | ||
| OPM2 | > 10 | 5 | GLO | ||
| PA-1 | > 10 | 7 | GLO | ||
| RPMI 8226 | > 10 | 5 | GLO | ||
| RS4;11 | > 10 | 5 | GLO | ||
| SK-BR-3 | > 10 | 5 | GLO | ||
| SK-N-MC | > 10 | 7 | GLO | ||
| TC-71 | > 10 | 7 | GLO | ||
| THP-1 | > 10 | 5 | GLO | ||
| U-2 OS | > 10 | 7 | GLO |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information3 | ID# |
| ACC-LC-319 | 4 – 6 | 2 | WST-8 | ||
| C6 | 1 – 5 | 3 | MTT | ||
| HeLa | 4 – 16 | 2 | MTT | G9a IC50 = 1.7 µM, GLP IC50 = 38 µM | |
| HeLa | < 5 | 2 | TBE | ||
| MCF7 | > 2 | 3 | HCC | ↓ Sox2 | |
| MCF7 | > 2 | 4 | HCC | ↓ Sox2 | |
| NCI-H2170 | > 6 | 2 | WST-8 | ||
| SBC-5 | < 2 | 2 | WST-8 | ||
| SHEP-1 | < 5 | 2 | TBE | ||
| SW780 | 2 – 4 | 2 | WST-8 | ||
| U-2 OS | < 5 | 2 | TBE | ||
| UM-UC-3 | 2 – 4 | 2 | WST-8 |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information | ID# |
| HeLa | > 13.3 | 2 | MTT |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information | ID# |
| HeLa | ~ 10 | 3 | GLO | ||
| HeLa | > 10 | 3 | CAA | ||
| HPAC | ~ 1 | 3 | GLO | ||
| HPAC | > 10 | 3 | CAA | ||
| MCF7 | 2.5 | 3 | GLO | ||
| MCF7 | 5 | 3 | CAA | ||
| PANC-1 | ~ 10 | 3 | GLO | ||
| PANC-1 | > 10 | 3 | CAA | ||
| PC-3 | ~ 10 | 3 | GLO | ||
| PC-3 | > 10 | 3 | CAA |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information3 | ID# |
| A-549 | 0.14 | 1 | SRB | ↓ SUV39H1, ↑ ATF3, CHOP | |
| AML-193 | 0.05 – 0.1 | 3 | MTT | Sub-G1 arrest (same by SUV39H1 shRNA); ↓ H3K9me2/3 on p15, CDH1 | |
| C6 | 0.01 – 0.05 | 3 | MTT | ||
| Calu-1 | 0.15 | 1 | SRB | ||
| CML-LSC | ~ 0.3 | 2 | TBE | ||
| HL-60 | ~ 0.1 | 1 | A5A | ↓ SUV39H1 & H3K9me3 on p15, CDH1, FZD9 | |
| K-562 | ~ 0.1 | 1 | A5A | ↓ SUV39H1 & H3K9me3 on CDH1, FZD9 | |
| Kasumi-1 | < 0.1 | 1 | A5A | ↓ SUV39H1 & H3K9me3 on p15, CDH1, FZD9 | |
| KG-1 | ~ 0.1 | 1 | A5A | ↓ SUV39H1 & H3K9me3 on p15, CDH1, FZD9 | |
| NCI-H157 | 0.1 | 1 | SRB | ↓ SUV39H1, ↑ ATF3, CHOP | |
| NCI-H1650 | 0.19 | 1 | SRB | ||
| NCI-H1792 | 0.22 | 1 | SRB | ↓ SUV39H1, ↑ ATF3, CHOP | |
| NCI-H460 | 0.15 | 1 | SRB | ||
| THP-1 | < 0.1 | 1 | A5A | ↓ SUV39H1 & H3K9me3 on CDH1, FZD9 |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information3 | ID# |
| A-549 | ~ 20 | 3 | MTT | ||
| DMS53 | < 5 | 5 | WST-8 | ||
| HepG2 | > 20 | 3 | MTT | ||
| HL-60 | ~ 1 | 2 | TBE | ↓ EZH2, SUZ12, Cyclin E | |
| HL-60 | 0.2 – 0.5 | 3 | A5A | ↓ EZH2, SUZ12, Cyclin E | |
| Jurkat | 12 | 2 | MTT | Cell cycle S-phase arrest | |
| Lu130 | ~ 5 | 6 | WST-8 | ||
| MCF10A | > 100 | 6 | MTS | ||
| MCF7 | 9.84 | 2 | MTT | ↓ EZH2 & DNMT1, ↑ RASSF2A | |
| MCF7 | 0.2 | 6 | MTS | G2/M arrest | |
| MDA-MB-231 | > 20 | 3 | MTT | ||
| MDA-MB-231 | 0.24 | 6 | MTS | G2/M arrest | |
| MPNST-724 | ~ 10 | 3 | MTT | ||
| NCI-H209 | < 5 | 6 | WST-8 | ||
| OCI-AML3 | ~ 0.5 | 2 | TBE | ↓ EZH2, SUZ12, Cyclin E | |
| OCI-AML3 | 0.2 – 0.5 | 3 | A5A | ↓ EZH2, SUZ12, Cyclin E | |
| S462 | 1 – 2 | 3 | MTT | G2 arrest | |
| SAS | < 10 | 2 | WST-8 | ||
| SK-BR-3 | ~ 0.8 | 6 | MTS |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information3 | ID# |
| DB | 0.37 – 1.1 | 15 | ACC | ||
| G-401 | ~ 1.1 | 15 | ACC | ↓ global H3K27me2/3 | |
| KARPAS 422 | 0.37 – 1.1 | 15 | ACC | ||
| OCI-Ly19 | > 10 | 10 | CFA | ↓ global H3K27me2/3 | |
| SU-DHL-4 | 3.3 – 10 | 15 | ACC | ||
| Toledo | 3.3 – 10 | 15 | ACC | ||
| WSU-DLCL2 | < 1 | 10 | CFA |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information | ID# |
| 697 | 36.57 | 14 | GVA | ||
| HL-60 | > 50 | 14 | GVA | ||
| Jurkat | > 50 | 14 | GVA | ||
| Kasumi-1 | 33 | 14 | GVA | ||
| KOPN-8 | 0.62 | 14 | GVA | ||
| MOLM-13 | 0.72 | 14 | GVA | ||
| MV4-11 | 0.17 | 14 | GVA | ||
| Reh | 13.9 | 14 | GVA | ||
| RS4;11 | 6.47 | 14 | GVA | ||
| SEM | 1.72 | 14 | GVA | ||
| THP-1 | 3.36 | 18 | GVA | ||
| U937 | > 50 | 14 | GVA |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information | ID# |
| Aska-SS | 0.72 | 14 | MTT | ||
| DoHH2 | 3.8 | 11 | GVA | ||
| Fuji | 1.5 | 14 | MTT | ||
| HT | 3 | 11 | GVA | ||
| KARPAS 422 | 0.45 | 11 | GVA | ||
| OCI-Ly19 | 1.7 | 11 | GVA | ||
| Pfeiffer | 0.005 | 11 | GVA | ||
| SU-DHL-6 | 0.32 | 11 | GVA | ||
| SYO-1 | 2.1 | 14 | MTT | ||
| Toledo | 13.4 | 11 | GVA | ||
| WSU-DLCL2 | 0.3 | 11 | GVA | ||
| Yamato-SS | 3.5 | 14 | MTT |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information | ID# |
| WSU-DLCL2 | 0.002-0.01 | 11 | GVA | Erradicates tumor in mice at 250 mg/kg |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information | ID# |
| Granta-519 | 0.06 | 12 | GVA | ||
| Jeko-1 | 0.9 | 12 | GVA | ||
| Maver-1 | 0.45 | 12 | GVA | | |
| Mino | 0.1 | 12 | GVA | ||
| Z-138 | 0.1 | 12 | GVA |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information3 | ID# |
| 697 | 32.7 | 14 | GVA | ||
| HL-60 | > 50 | 14 | GVA | ||
| Jurkat | > 50 | 14 | GVA | ||
| Kasumi-1 | 12.2 | 14 | GVA | ||
| KOPN-8 | 0.17 | 14 | GVA | ||
| MOLM-13 | 0.06 | 14 | GVA | ||
| MOLM-13 | > 1 | 7 | A5A | ↓ HOXA9, MEIS1 | |
| MOLM-13 | ~ 0.16 | 7 | A5A | ↓ HOXA9, MEIS1 | |
| MV4-11 | 0.004 | 14 | GVA | Sub-G1 arrest | |
| NOMO-1 | 0.03 | 14 | GVA | ||
| Reh | 1.2 | 14 | GVA | ||
| RS4;11 | 1.3 | 14 | GVA | ||
| SEM | 0.2 | 14 | GVA | ||
| SKM-1 | > 4 | 7 | GLO |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information3 | ID# |
| Fuji | 0.15 | 14 | GLO | ↑ ATF3 | |
| Fuji | 0.15 | 11 | CAA | ↑ ATF3 | |
| G-401 | 0.16 | 14 | GLO | ||
| HS-SY-2 | 0.52 | 14 | GLO | ↑ CDKN2A | |
| HS-SY-2 | 0.63 | 11 | GLO | ↑ CDKN2A | |
| SW982 | > 10 | 14 | GLO |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information | ID# |
| 697 | ~ 7.5 | 6 | GLO | ||
| BC-1 | 8.29 | 6 | GLO | ||
| BC-2 | 4.76 | 6 | GLO | ||
| BC-3 | 2.22 | 6 | GLO | ||
| CA46 | 6.59 | 6 | GLO | ||
| Ci-1 | 4.28 | 6 | GLO | ||
| COLO 205 | ~ 5 | 3 | MTT | ||
| CRO-AP2 | 1.64 | 6 | GLO | ||
| Daudi | 1.27 | 6 | GLO | ||
| DB | 0.86 | 6 | GLO | ||
| DB | 0.25 | 9 | ACC | ||
| DG-75 | 3.25 | 6 | GLO | ||
| DMS53 | ~ 8 | 8 | WST-8 | ||
| Farage | 1.72 | 6 | GLO | ||
| HCT 116 | ~ 5 | 3 | MTT | ||
| HD-MY-Z | 10.72 | 6 | GLO | ||
| Hs 445 | 3.53 | 6 | GLO | ||
| HS-Sultan | 2.28 | 6 | GLO | ||
| HT | 0.52 | 6 | GLO | ||
| HT-29 | 2 – 5 | 3 | MTT | ||
| Jiyoye | 0.23 | 6 | GLO | ||
| KARPAS 231 | ~ 3 | 6 | GLO | ||
| KARPAS 422 | 0.23 | 6 | GLO | ||
| KARPAS 422 | > 3 | 7 | GLO | ||
| KARPAS 422 | ~ 3 | 8 | GLO | ||
| KARPAS-1106P | 4.54 | 6 | GLO | ||
| L-428 | 4.7 | 6 | GLO | ||
| LNCaP | > 10 | 2 | PIA | ||
| Lu130 | ~ 8 | 8 | WST-8 | ||
| MC116 | 10.17 | 6 | GLO | ||
| MHH-PREB-1 | 5.3 | 6 | GLO | ||
| Mino | 7.34 | 6 | GLO | ||
| MLL-AF9 | 3 – 10 | 7 | GLO | ||
| MLL-AF9 | > 3 | 12 | ACC | ||
| MN60 | ~ 3.5 | 6 | GLO | ||
| Myc-CaP/AS | < 10 | 2 | PIA | ||
| NALM-6 | 2.5 | 6 | GLO | ||
| NALM-6 | ~ 3.13 | 5 | CAA | ||
| NCI-H209 | < 8 | 8 | WST-8 | ||
| NU-DUL-1 | 17.06 | 6 | GLO | ||
| OCI-Ly19 | < 10 | 4 | GLO | ||
| OCI-Ly19 | 1.02 | 6 | GLO | ||
| OCI-Ly19 | ~ 3 | 12 | GLO | ||
| OVISE | < 5 | 12 | A5A | ||
| P3HR-1 | 3.21 | 6 | GLO | ||
| Pfeiffer | 0.03 | 6 | GLO | ||
| Raji | 7.87 | 6 | GLO | ||
| RC-K8 | 4.53 | 6 | GLO | ||
| Reh | ~ 4 | 6 | GLO | ||
| Ri-1 | 7.66 | 6 | GLO | ||
| RL | 4.73 | 6 | GLO | ||
| RMG-1 | > 5 | 12 | A5A | ||
| RPMI-6666 | 1.43 | 6 | GLO | ||
| SC-1 | 3.73 | 6 | GLO | ||
| SU-DHL-4 | 4.83 | 6 | GLO | ||
| SU-DHL-5 | 2.3 | 6 | GLO | ||
| SU-DHL-6 | 0.58 | 6 | GLO | ||
| SU-DHL-8 | 3.19 | 6 | GLO | ||
| SU-DHL-10 | 0.45 | 6 | GLO | ||
| SU-DHL-16 | 3.28 | 6 | GLO | ||
| SUP-B8 | ~ 0.4 | 6 | GLO | ||
| SUP-B8 | ~ 0.78 | 5 | CAA | ||
| SUP-B8 | ~ 0.1 | 6 | CAA | ||
| SUP-B15 | ~ 7 | 6 | GLO | ||
| TANOUE | ~ 5 | 6 | GLO | ||
| Toledo | 13.79 | 6 | GLO | ||
| TOV-21G | ~ 0.27 | 12 | A5A | ||
| TRAMP-C2 | < 10 | 2 | PIA | ||
| U-2932 | 2.94 | 6 | GLO | ||
| U-2940 | 4.56 | 6 | GLO | ||
| U-698-M | 5.6 | 6 | GLO | ||
| WERI-Rb-1 | 2.5 – 5 | 2 | GLO | ||
| WILL-1 | 5.53 | 6 | GLO | ||
| WILL-2 | 27.46 | 6 | GLO | ||
| WSU-DLCL2 | 0.13 | 6 | GLO | ||
| WSU-FSCCL | 11.97 | 6 | GLO | ||
| WSU-NHL | 3.54 | 6 | GLO | ||
| Y-79 | 2.5 – 12.5 | 2 | GLO |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information3 | ID# |
| A-549 | ~ 15 | 3 | MTT | ||
| HepG2 | 15 – 20 | 3 | MTT | ||
| MDA-MB-231 | 10 – 15 | 3 | MTT | ||
| OVCAR-10 | > 1 | 12 | HCC | ↓ global H3K27me3 | |
| SK-OV-3 | > 1 | 12 | HCC | ↓ global H3K27me3 | |
| UPN289 | > 1 | 12 | HCC | ↓ global H3K27me3 |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information3 | ID# |
| KARPAS 422 | ~ 1 | 7 | GLO | ||
| KARPAS 422 | 3 – 10 | 4 | GLO | ||
| MLL-AF9 | ~ 3 | 7 | GLO | ↓ global H3K27me3 | |
| OCI-Ly19 | > 10 | 12 | GLO | ||
| WERI-Rb-1 | 5 – 12.5 | 2 | GLO | ||
| Y-79 | 5 – 12.5 | 2 | GLO |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information3 | ID# |
| 22Rv1 | 4.5 | 2 | MTT | ||
| BON-1 | ~ 2.5 | 2 | ACC | ↓ c-Myc, global H3K9me2; ↑ DKK-1&3, WIF-1 | |
| HCT116 | 11 | 2 | MTT | ||
| MCF7 | 7.6 | 2 | MTT | ||
| MDA-MB-231 | 11 | 2 | MTT | ||
| PC-3 | 14 | 2 | MTT |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information3 | ID# |
| MDA-MB-231 | > 1 | 14 | CFA | ||
| MDA-MB-231 | 16.7 | 2 | RCA | H3K9me2 IC50 = 110 nM | |
| PANC-1 | 1 | 14 | CFA | Selective for PRC2/EZH2 vs other HMTs & DNMT1 | |
| PANC-1 | 3.5 | 2 | RCA | H3K9me2 IC50 = 40 nM | |
| PC-3 | 8.9 | 2 | RCA | H3K9me2 IC50 = 130 nM | |
| U-2OS | 6 | 2 | RCA | H3K9me2 IC50 = 130 nM |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information3 | ID# |
| MDA-MB-231 | > 50 | 2 | RCA | ↓ global H3K9me2 |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.
| Cell line | IC50 (µM) | Inc1 | Assay2 | Additional information3 | ID# |
| DB | 0.63 | 8 | ACC | ||
| DB | 0.23 | 16 | ACC | ||
| EOL-1 | 0.1 | 16 | ACC | ||
| EOL-1 | ~ 0.5 | 8 | ACC | ||
| HT-29 | 5 | 3 | MTS | ||
| K-562 | > 250 | 16 | ACC | ||
| LOUCY | > 200 | 16 | ACC | ||
| MCF10A | 19.2 | 3 | RCA | ||
| m-MLL-AF9 | 0.4 | 16 | ACC | G0/G1 arrest; ↑ p16, p19 | |
| m-MLL-AF9 | ~ 3 | 8 | CFU | G0/G1 arrest; ↑ p16, p19 | |
| m-MLL-ENL | 0.55 | 16 | ACC | ||
| m-MLL-ENL | ~ 3 | 4 | ACC | ||
| m-MLL-ENL | < 3 | 8 | CFU | ||
| MV4-11 | 0.53 | 16 | ACC | ||
| OCI-AML2 | 1.5 | 16 | ACC | ||
| RS4;11 | 1.96 | 16 | ACC |
© CEDD, Cancer Epigenetics Society & B² Scientific Group, Ltd. All rights reserved, 2016. Data may be included in academic publications contingent on the citation of the reference on top of this page.